|
Selecting a Template for Data Import
Overview
GeneLinkerô can read expression data files produced by a wide variety of other software. GeneLinkerô uses a template to interpret the contents of your data file or files.
Data files containing one sample each:
|
Template Name |
Template Description |
|
Import Affymetrix MAS 4.0 data files. | |
|
Import Affymetrix MAS 5.0 data files. | |
|
Import CodeLink XML files. | |
|
Import dChip paired xls files. | |
|
Import GenePix ATF data files. | |
|
Import single channel or two color Koadarray files. | |
|
Import GenePix ATF two-color data values (treatment=green/control=red). | |
|
Import GenePix ATF data files and generate reliability measures by merging replicates (see Merging Within-Chip Replicate Measurements). | |
|
Import GenePix ATF two-color data values (treatment=red/control=green). | |
|
Import Genomic Solutions files. | |
|
Import Genomic Solutions data files and generate reliability measures by merging replicates (see Merging Within-Chip Replicate Measurements). | |
|
Import Quantarray data values into a two-color dataset. | |
|
Import ScanArray data files. | |
|
Import ScanArray data files and generate reliability measures by merging replicates (see Merging Within-Chip Replicate Measurements). | |
|
Import ScanArray two-color data values (treatment=Ch1/control=Ch2). | |
|
Import ScanArray two-color data values (treatment=Ch2/control=Ch1). | |
|
Import spectral (proteomics) data and peak files. |
Data files containing all samples in one file:
|
Multi-Sample Data File |
Template Description |
|
Import dChip single xls data file. | |
|
Import tabular data from a single multi-sample data file. | |
|
Import tabular data with genes represented by columns and generate reliability measures by merging replicate genes (see Merging Within-Chip Replicate Measurements). Be sure this is what you want. Tabular files more typically have genes in rows! | |
|
Import tabular data with genes represented by rows and generate reliability measure by merging replicate genes (see Merging Within-Chip Replicate Measurements). | |
|
If you have generated reliability measures for tabular data independently of GeneLinkerô, it is possible to import them along with your data. They must be in a tabular file of identical shape to your gene expression data file. If your gene expression data file is named FileName.ext then your reliability measures must be in a file named FileName_rm.ext in the same folder. GeneLinkerô expects that reliability measures will be between 0 and 1 inclusive, and that values close to 0 will indicate highly reliable data. |
If you do not see a format in the lists above that matches the format of your data, your best course of action is to transform your data into Tabular format. See Importing Data from Tabular Files for more information.
Actions
1. Click the Import
Gene Expression Data toolbar icon ![]() , or select Import from the File
menu and Gene Expression Data
from the sub menu. The Data Import
dialog is displayed.
, or select Import from the File
menu and Gene Expression Data
from the sub menu. The Data Import
dialog is displayed.
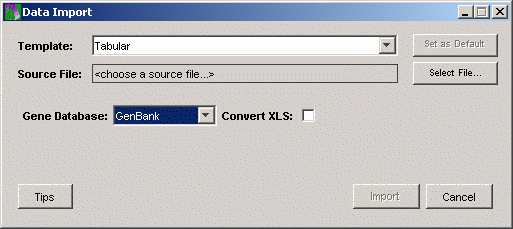
2. Select a Gene Database from the drop-down list. This tells GeneLinkerô which type of gene identifier the genes being imported have (GenBank, Affymetrix, UniGene, or custom). If you need more information about this, see Selecting the Gene Database Type. You can also select the gene database after you have changed templates if you wish.
The name of the selected template appears on the Data Import dialog. If this is the appropriate template for your data, go to either Importing One File Containing All Samples, or Importing Multiple Files With One Sample Each, as appropriate to the template you have selected.
If the appropriate template name is not showing on the dialog, then continue:
3. Click the template that is appropriate for your data file(s) from the drop-down list. The template is highlighted.
4. To set the selected template as the default, click the Set As Default button. If you will be importing data of the same format repeatedly, you should do this so you will not need to re-select the same template each time you import data. When this button is inactive (text is grey) the displayed template is the default template.
5. Click Select.
If you selected one of the Tabular or DCHIP single xls file templates, the Data Import dialog is updated to permit you to specify a single data file to import, which should contain data about all the samples.
Go to Importing One File Containing All Samples.
If you selected an Affymetrix, CodeLink, DCHIP paired xls files, GenePix, Genomic Solutions, Quantarray or ScanArray template, the Data Import dialog layout changes to permit you to select a set of single-sample data files from one folder.
Go to Importing Multiple Files With One Sample Each.
Note: gene identifiers have a length restriction of 25 characters. This means that on import of a dataset or a gene list, identifiers that are longer than 25 characters are truncated.
Related Topics:
Importing One File Containing All Samples
Importing Multiple Files With One Sample Each
Merging Within-Chip Replicate Measurements

