|
Importing Data from Quantarray Files
Overview
The data files must be in the Quantarray file format.
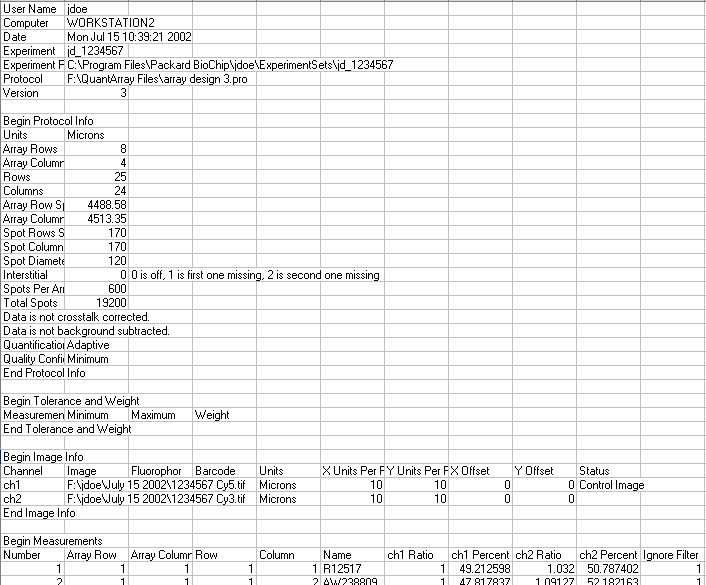
File Header Section Example:
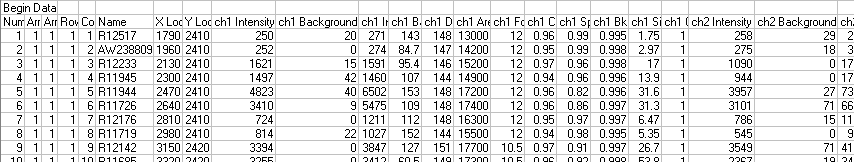
File Data Section Example:
Import Process
Multiple files are processed into a single two-color dataset. The sample order of the imported dataset is determined by the order of the source sample data files listed in the Import Data dialog.
Characteristics of the Quantarray Import Template
The Quantarray import template assumes the following about the format of the data files:
1. The data must be delimited by tab characters.
2. Gene identifiers are in the sixth column of the Data section.
3. The Measurements section is ignored.
4. Treatment and control channels are based on the information in the Image Info section of the Quantarray files. NOTE: All files must use the same channel (either ch1 or ch2) for the control channel. The channel used for control in all files is the channel labelled 'Control Image' in the last file in the import list. You can reorder the files in the import list using the black up- and down-arrow buttons on the Data Import dialog.
5. If the Image Info section is missing from the last file, then ch1 is used for the control channel and ch2 for the treatment channel.
6. It is assumed that the foreground and background counts are found in the Data section in the columns headed ch1 Intensity, ch1 Background, ch2 Intensity and ch2 Background. The substrings 'ch1' and 'ch2' must match the lines in the Image Info section, if present.
7. GeneLinkerô stores the resulting ratios and associated intensities in a two-color dataset listed in the navigator. This makes it possible (for instance) to apply a Lowess correction to the dataset.
8. Spots for which the background count exceeds the foreground count are imported into GeneLinkerô as missing values. Negative ratios are not imported.
Related Topics:
Selecting a Template for Data Import
Importing Multiple Files With One Sample Each

