|
Variables Overview
Overview
Definition of a Variable
In GeneLinkerô, a variable is a column of data other than gene expression values used to differentiate samples. A variable can store:
Phenotypic observations about the samples.
e.g. malignant vs. benign.
Predictions of phenotypes by a trained classifier.
e.g. predicted malignant vs. predicted benign.
Information about experimental conditions.
e.g. high dose vs. low dose; time the sample was taken; animal A vs. animal B vs. animal C, etc.
Variable File Formats
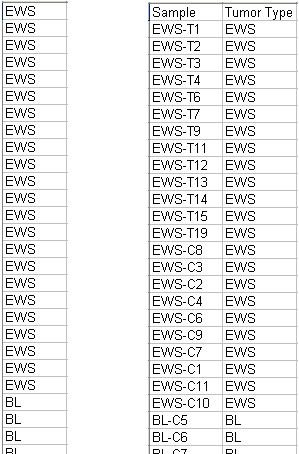
One-column: A one-column format file consists of the class name of each sample, one per line, in the same sample order as in the expression data file. The first row must not contain a column header.
Two-column: The two-column format has the sample names in the first column and the variable values (class names) in the second. The two-column format can be tab-separated or comma-separated. If you want class names which include commas, you must use two-column format with tab separators between the sample names and class labels. The first row must contain column headers.
Uses of a Variable
Variables can be used many ways in GeneLinkerô.
You can color the samples in certain plots by a variable.
A variable can group replicates together for statistical differentiation using the F-Test. All members of the same group have the same variable value.
SLAMô can search for gene sets associated with the values of a variable.
A variable can be used as training data for an ANN classifier or an IBIS classifier and a trained classifier can predict the values of a variable for new samples.
Two variables of the same type can be compared using a confusion matrix.
Note on the Value 'Unknown'
Any GeneLinkerô variable may take on the special value of 'Unknown'. In the output of a trained classifier, this means that the classifier could not make a reliable prediction of the sample class. In other contexts, 'Unknown' is treated in the same manner as any other class. To reduce confusion we recommend that you use more informative class labels and reserve 'Unknown' for the output of the classifier.
Variable Types
Variables which attempt to describe the same phenomenon are grouped together into a Variable Type. GeneLinkerô does not intuit which variables refer to the same phenomenon the way a person does, so you must define a variable type for each variable you import.
For example, variables of type 'leukemia class' might have possible values of 'myeloblastic' and 'lymphoblastic'. Once you have created the variable type 'leukemia class', you could then import variables of that type like 'Diagnosis of pathologist A', 'Diagnosis of pathologist B', etc. You could then go on to train GeneLinkerô to classify the samples by leukemia type, and use GeneLinkerô to construct further variables like 'Prediction based on gene Q', 'Prediction based on a set of 10 genes', and so on.
If you wished to study disease outcomes with the same expression dataset, you could define a new variable type 'outcome' which might have values such as 'survived' and 'died'. You could then import a variable of that type, train classifiers and attempt further predictions.
Observed vs. Predicted Variables
In GeneLinkerô, imported variables are referred to as observed variables, and variables generated by a classifier are predicted. You can see the values of any or all of the variables associated with a given dataset using the Variable Viewer. You can edit, delete, compare or export variables using the Variable Manager.
Variable Indicator
In the Experiments navigator, a root dataset that has one or more variables associated with it has the variables tag on the icon next to its name. The same variables are associated with all the descendants of this dataset.
![]() for a complete dataset.
for a complete dataset.
![]() for an incomplete dataset.
for an incomplete dataset.
Variables and Classification
Variables are typically imported into GeneLinkerô for one of two purposes related to Classification: A variable may be a training target, providing known classes for training a classifier, or a variable may be a set of test results for comparison with the predictions of a trained classifier. Note that for a given prediction problem, both the training variable and the test variable must be imported as the same Variable Type.
Related Topics:
Variables in Supervised Learning

