|
Platinum
SLAMô
Overview
SLAMô (Sub-Linear Association Mining) is a proprietary data mining algorithm of Predictive Patterns Software Inc. (PPSI) that is used to find correlations between discretized variables or to predict the outcome of a categorical variable. As an aid to supervised learning, SLAMô is used to find associations in gene expression data so that a list of interesting genes (features) can be created.
Association Mining Overview
Association mining is a machine learning technique which detects when sets of variables have certain values occuring together at a rate greater than would happen by chance. In GeneLinkerô, the variables are genes. SLAMô finds sets of gene expression values which co-occur frequently within each dataset. Such sets are called associations. For instance, it may happen that in kidney tissue, repression of gene A results in the up-regulation of genes B and C, and down-regulation of gene Q. In this case, we would expect to find an association like this in the dataset :
Kidney Tissue: Gene A: low, gene B: high, gene C: high, gene Q: low.
Note: this says nothing about how B, C, and Q are regulated when A is not repressed, or when a different tissue is being considered.
Such an association can be used in GeneLinkerô to find genes which are connected to certain sample classes. Genes which occur in many such associations, or in associations with very high support (see below), are likely to be good predictors Ė that is to say, good candidates for classification features.
Association Statistics
Support: the support statistic of an association is the number of samples in the dataset in which that association appears.
Matthews correlation: a measure of the predictive power of an association: How well those gene values predict that particular class. (Note that this is not related in any simple fashion to the ability of those same genes to predict other classes.)
Actions
1. Click a Discretization item in the Experiments navigator. The item is highlighted.
2. Click the SLAM
toolbar icon ![]() , or select SLAM
from the Predict menu, or right-click
the item and select SLAM from
the shortcut menu. The SLAM parameters
dialog is displayed.
, or select SLAM
from the Predict menu, or right-click
the item and select SLAM from
the shortcut menu. The SLAM parameters
dialog is displayed.
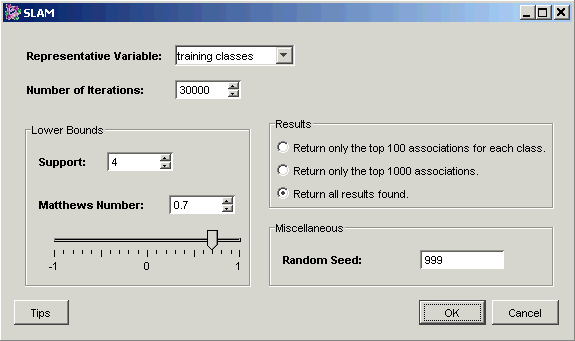
3. Set the parameters.
|
Parameter |
Description |
|
Representative Variable |
The training variables to be used for prediction |
|
Number of Iterations |
The number of SLAMô iterations. |
|
Support Lower Bound |
Minimum support threshold for SLAMô. |
|
Matthews Number Lower Bound |
Minimum Matthews threshold for SLAMô. |
|
Results |
If the Matthews and Support bounds settings result in a large number of valid associations being discovered, this setting can be used to limit the results returned to the best 100 or 1000 associations. |
|
Random Seed |
The seed value for the random number generator. In normal use, setting the random seed is neither necessary nor recommended. On occasion, you may need to determine whether a certain variation in results is due to the random element, or some other cause. For this reason, you are able to set the random seed to a fixed value, thus controlling that source of variation. In SLAMô, the random seed can be thought of as prescribing the starting point for the search for associations. If SLAMô is allowed to run long enough, it will find all of an enormous set of associations which inhabit any given dataset, but the smaller you set the number of iterations, the greater will be the effect of the random seed. Conversely, the random seed matters less and less as the number of iterations grows greater. It is usually better to set the iteration number high and let SLAMô run overnight than to do repeated runs with different random seeds. |
4. Click OK. The Experiment Progress dialog is displayed. It is dynamically updated as the SLAM operation is performed. To cancel the SLAM operation, click the Cancel button.
Upon successful completion, a new item (SLAM) is added under the Discretization item in the Experiments navigator.
If automatic visualizations are enabled in your user preferences, the SLAM Association Viewer is displayed upon completion of the SLAM run.
Related Topics:
ANN Classification and Prediction Overview

