|
Platinum
Tutorial 7: Step 6 Perform IBIS 2D LDA Search
Overview
Perform an IBIS 2-dimensional search over gene pairs. A 2-dimensional search takes longer than the 1-dimensional search performed previously. IBIS can examine every possible pair of genes in the dataset (1041 * 1040 / 2 = 541320 pairs) and evaluate the MSE and accuracy of each classifier (gene pair) on that data.
For the purposes of this tutorial, we will use the 1D IBIS results to filter down the number of genes that will be searched by 2D IBIS. However, if we were to simply choose the best 1D classification genes, we would expect that two-dimensional combinations of them would also produce fairly good classification just because the individual genes were already fairly good. So instead we shall use 2D IBIS to examine the genes that are not good 1D predictors, to see if there are cases where combinatorial effects are prominent.
Actions
1. Click the IBIS Search Results: 1D LDA window to bring it to the front. If you closed the window, double-click on the Thiopurine IBIS search results item in the Experiments navigator. The IBIS Results Viewer is displayed.
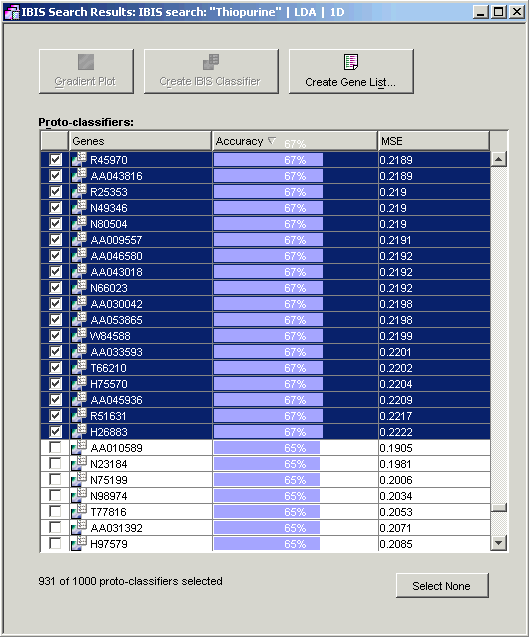
2. Ensure that the results are sorted by Accuracy (the default).
3. Click the checkbox to the left of the top gene (AA046755) so that it is checked.
4. Scroll down until accuracy values of 67% and 65% are visible. Press and hold the <Shift> key and click the checkbox to the left of the last gene with an accuracy of 67% (H26883). This checks every gene from the top gene down to this one.
5 Click the Create Gene List button. The Create a Gene List dialog is displayed.
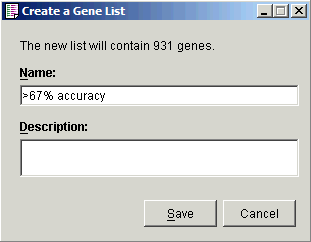
6. For the Name, type in >67% accuracy.
7. Click Save. A gene list is created and added to the Gene Lists navigator.
7. Click the NCI60 basal expression dataset item in the Experiments navigator. The item is highlighted.
8. Click the Filter
Genes toolbar icon![]() , or select Filter
Genes from the Data menu,
or right-click the item and select Filter
Genes from the shortcut menu. The Filter
Genes dialog is displayed.
, or select Filter
Genes from the Data menu,
or right-click the item and select Filter
Genes from the shortcut menu. The Filter
Genes dialog is displayed.
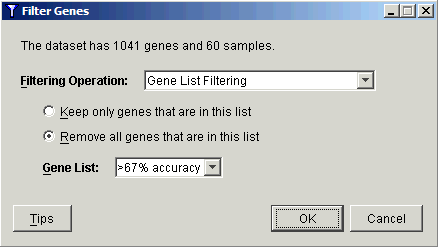
9. Select Gene List Filtering from the Filtering Operation drop-down list.
10. Select Remove all genes that are in this list.
11. Select the gene list > 67% accuracy from the Gene List drop-down list.
12. Click OK. A new Filtered: removed {> 67% accuracy} dataset is added to the Experiments navigator. It contains the 110 genes which had less than 67% accuracy as 1D linear predictors of thiopurine response.
13. If the new Filtered: removed {> 67% accuracy} dataset in the Experiments navigator is not already highlighted, click it.
14. Click the IBIS
Classifier Search toolbar icon ![]() , or select IBIS Classifier Search from the Predict menu, or right-click the item
and select IBIS Classifier Search
from the shortcut menu. The menu. The IBIS
Classifier Search dialog is displayed.
, or select IBIS Classifier Search from the Predict menu, or right-click the item
and select IBIS Classifier Search
from the shortcut menu. The menu. The IBIS
Classifier Search dialog is displayed.
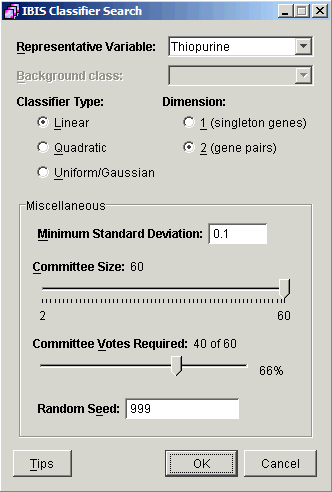
3. Set the parameters.
|
Parameter |
Setting |
|
Representative Variable |
Thiopurine |
|
Classifier Type |
Linear |
|
Dimension |
2 (gene pairs) |
|
Minimum Standard Deviation |
0.1 |
|
Committee Size |
60 |
|
Committee Votes Required |
40 of 60 (66%) |
|
Random Seed |
999 |
4. Click OK. The IBIS 2D LDA search is performed and a new item IBIS Search Results LDA 2D is added to the Experiments navigator under the original dataset. This typically takes 5 to 10 minutes depending on the speed and load of your machine.
If you have automatic visualization enabled in your user preferences, the IBIS Search Results Viewer is displayed.

