|
Tutorial 1: Step 6 Perform Partitional Clustering
From visual examination of a hierarchical clustering, Wen et al. identified five groups or 'waves' plus a small number of outliers or 'other' genes. This step will demonstrate that GeneLinkerô can be used to get a similar clustering, using the K-Means clustering function.
The key feature of K-Means clustering is that you choose a priori the number of clusters you think the data should be divided into. This number is the 'K' in K-Means.
The K-Means algorithm uses the same Euclidean Average-Linkage distance metric used for hierarchical clustering earlier.
Perform Partitional Clustering
1. If the renamed normalization item in the Experiments navigator is not already highlighted, click it.
2. Click the Partitional
Clustering toolbar icon ![]() , or select Partitional
Clustering from the Clustering
menu, or right-click the item and select Partitional
Clustering from the shortcut menu. The Partitional
Clustering parameters dialog is displayed.
, or select Partitional
Clustering from the Clustering
menu, or right-click the item and select Partitional
Clustering from the shortcut menu. The Partitional
Clustering parameters dialog is displayed.
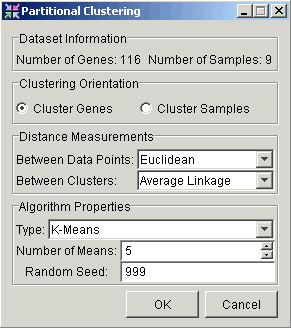
3. Set dialog parameters.
|
Parameter |
Setting |
|
Clustering Orientation |
Cluster Genes |
|
Distance Measurements: Between Data Points |
Euclidean |
|
Distance Measurements: Between Clusters |
Average Linkage |
|
Algorithm Properties: Type |
K-Means |
|
Algorithm Properties: Number of Means |
5 |
|
Algorithm Properties: Random Seed |
999 |
3. Click OK. The clustering operation is performed and upon successful completion, a new Gene Partitional Clustering experiment is added to the Experiments navigator under the original dataset. Rename it if you like.
If you have automatic visualizations enabled in your user preferences, a matrix tree plot of the clustering results is displayed. You can close this plot when you are finished looking at it.
Use of the Random Seed Parameter
In normal use, setting the random seed is neither necessary nor recommended. In a tutorial you set the random seed to a consistent value so that you will obtain precisely the same results that we depict and discuss, which makes the tutorial easier to understand. When you are not following a tutorial, you should generally not adjust the random seed at all.
The random seed setting may affect irrelevant details, such as the labelling and ordering of clusters. In other cases the random seed may affect relevant details, such as which genes occur together in clusters. Because of this latter possibility, it is sometimes worth repeating an experiment with different random seeds to see what the effects are. (In step 7 see 'The Centroid Plot: Variability in K-Means Clustering' below.) GeneLinkerô helps with this by setting a new random seed every time an operation is carried out, so you donít need to.
On occasion you may need to determine whether a certain variation in results is due to the random element, or some other cause. For this reason you are able to set the random seed to a fixed value, thus controlling that source of variation.

