|
Tutorial 1: Step 4 Perform Hierarchical Clustering
In this step of the tutorial, you will perform a hierarchical clustering experiment on the normalized data to reveal its intrinsic structure. For complete details on the clustering operations available in GeneLinkerô, please see Clustering Overview.
Perform Hierarchical Clustering:
1. If the renamed normalization dataset in the Experiments navigator is not already highlighted, click it.
2. Click the Hierarchical
Clustering toolbar icon ![]() , or select Hierarchical
Clustering from the Clustering
menu, or right-click the item and select Hierarchical
Clustering from the shortcut menu. The Hierarchical
Clustering parameters dialog is displayed.
, or select Hierarchical
Clustering from the Clustering
menu, or right-click the item and select Hierarchical
Clustering from the shortcut menu. The Hierarchical
Clustering parameters dialog is displayed.
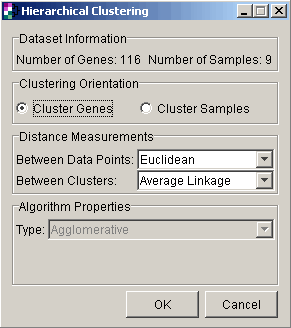
3. Set dialog parameters.
|
Parameter |
Setting |
|
Clustering Orientation |
Cluster Genes |
|
Distance Measurement: Between Data Points |
Euclidean |
|
Distance Measurement: Between Clusters |
Average Linkage |
4. Click OK. The clustering operation is performed and upon successful completion, a new Gene Hierarchical Clustering experiment is added to the Experiments navigator under the normalized dataset. You can rename it if you wish.
If you have automatic visualizations enabled in your user preferences, a matrix tree plot of the clustering results is displayed.

