|
Tutorial 5: Step 2 Principal Component Analysis
Principal Component Analysis
1. If the Elutriation dataset in the Experiments navigator is not already highlighted, click it.
2. Click the Principal
Component Analysis toolbar icon ![]() , or select Principal
Component Analysis from the PCA
menu, or right-click the item and select Principal
Component Analysis from the shortcut menu. The PCA
parameters dialog is displayed.
, or select Principal
Component Analysis from the PCA
menu, or right-click the item and select Principal
Component Analysis from the shortcut menu. The PCA
parameters dialog is displayed.
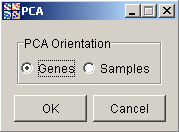
3. You may choose to perform PCA calculation on either Genes or Samples. The typical use of PCA is to reduce the genes to a smaller number of 'variables' as in this tutorial. Ensure that Genes is selected. (In other applications, where the samples are being thought of as 'variables' or measurements for particular genes, you would select Samples).
4. Click OK. The Experiment Progress dialog is displayed.
The dialog is dynamically updated as the PCA calculation is performed. Upon successful completion, a PCA: genes item is added to the Experiments navigator under the original dataset.
If you have automatic visualizations enabled in your user preferences, a 3D Score Plot is displayed.

