|
Tutorial 2: Step 6 Perform Hierarchical Clustering
Perform Hierarchical Clustering
1. Click the 3-nearest neighbors dataset in the Experiments navigator (Click the Experiments tab to display the Experiments navigator). The item is highlighted.
2. Click the Hierarchical
Clustering toolbar icon ![]() , or select Hierarchical
Clustering from the Clustering
menu, or right-click the item and select Hierarchical
Clustering from the shortcut menu. The Hierarchical
Clustering dialog is displayed.
, or select Hierarchical
Clustering from the Clustering
menu, or right-click the item and select Hierarchical
Clustering from the shortcut menu. The Hierarchical
Clustering dialog is displayed.
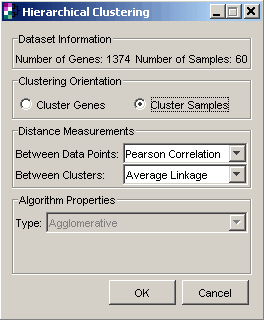
3. Set parameters.
|
Parameter |
Setting |
|
Clustering Orientation |
Cluster Samples |
|
Data Measurements: Between Data Points |
Pearson Correlation |
|
Data Measurements: Between Clusters |
Average Linkage |
Note that Agglomerative (the default option) is set as the Type parameter in the Algorithm Properties group.
4. Click OK. The clustering operation is performed, and upon successful completion, a new Sample Hierarchical Clustering experiment is added to the Experiments navigator under the original dataset.
GeneLinkerô provides many different clustering algorithms, and there are other clustering methods listed under Partitional Clustering. Genes can be clustered in addition to samples by using the same command sequence as above but changing the choice of clustering orientation from Samples to Genes.
If you have automatic visualizations enabled in your user preferences, a matrix tree plot of the clustering results is displayed.

