|
Removing Values by Reliability Measure
Overview
This function is used to create missing values from unreliable gene expression values. Unreliability might be implied by a poor reliability measurement, or it might be implied by an extreme expression measurement.
This function can only be applied to top-level datasets that have associated reliability measurement data. The reliability measure may be a P-Value imported from an Affymetrix MAS 5.0 file, or computed on import from within-chip replicates.
The result of this operation can be either a complete or an incomplete dataset.
Actions
1. Click a complete or incomplete dataset with associated reliability measures in the Experiments navigator. The item is highlighted.
2. Select Remove Values from the Data menu, or right-click the item and select Remove Values from the shortcut menu. The Remove Values parameters dialog is displayed.
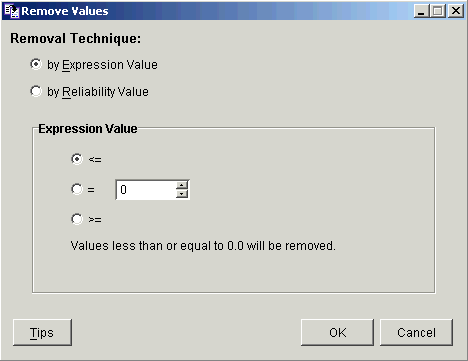
3. Click by Reliability Value as the Removal Technique. The dialog is updated.
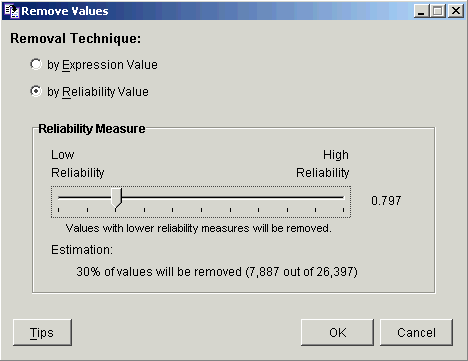
4. Use the slider to set the reliability measure threshold. The reliability scale is from 1.0 (low reliability) to 0.0 (high reliability).
5. Click OK. The Experiment Progress dialog is displayed. It is dynamically updated as the value removal operation is performed.
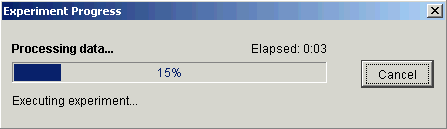
Upon successful completion, a new dataset is added under the original dataset item in the Experiments navigator.
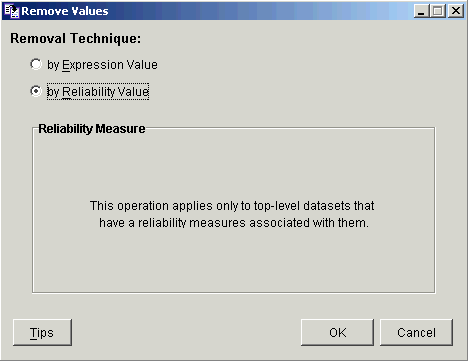
If the dataset you selected is not a top-level dataset, or if it does not have reliability data associated with it, the dialog is updated to indicate this. Click OK to exit this operation.
Related Topic:
Creating a Table View of Reliability Data

