|
Platinum
Tutorial 6: Step 3 Discretize the Data
The first step in our analysis of this dataset is to use SLAMô to look for associations between multiple genes and the tumor type.
SLAMô finds associations between genes based on identical patterns of gene expression. For example, if Gene A is HIGH whenever Gene B is LOW, SLAMô identifies an association between Gene A and Gene B. Because the number of possible patterns is enormous, particularly when looking for patterns between five or ten genes rather than just two, we need a fast, simple means of comparing expression levels. By discretizing the data, it becomes possible to compare expression levels in terms of a small number of discrete categories (e.g. HIGH/MEDIUM/LOW) rather than continuous values. This speeds up the comparison process by many orders of magnitude.
Discretize the Data
1. Click the Khan_training_data dataset in the Experiments navigator. The item is highlighted.
2. Click the Discretize
toolbar icon ![]() , or select Discretize
Data from the Predict menu,
or right-click the item and select Discretize
Data from the shortcut menu. The Discretization
parameters dialog is displayed.
, or select Discretize
Data from the Predict menu,
or right-click the item and select Discretize
Data from the shortcut menu. The Discretization
parameters dialog is displayed.
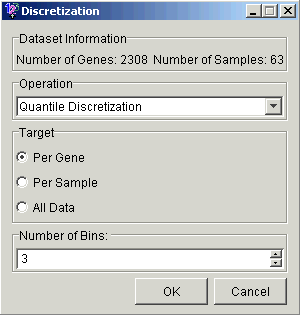
Operation Type
Quantile Discretization means dividing the data into equally-populated groups. Thus 3-way quantile discretization per gene will yield a roughly equal number of high (2), medium (1) and low (0) values for each gene.
Range Discretization makes the groups cover equal ranges. For example, if the gene had values ranging from 0.0 to 24.0, a 3-way range discretization would consist of values between 0 and 8, 8 and 16, and 16 and 24, and the three groups might be quite differently populated.
Number of Bins
Choosing the number of bins is a balancing act. The more bins you use, the less information is discarded by the discretization. But the more bins there are, the fewer associations SLAMô will find.
Accept the default parameters (Quantile discretization, Per Gene, and 3 bins).
3. Click OK. The Experiment Progress dialog is displayed. It is dynamically updated as the discretization operation is performed.
Upon successful completion, a new Discretized: 3 bins/gene | quantile dataset is added under Khan_training_data in the Experiments navigator.

